Python How to Read Data From an Excel
Making Sense of Big Data
Do Y'all Read Excel Files with Python? In that location is a 1000x Faster Way.
In this article, I'll show y'all 5 ways to load data in Python. Achieving a speedup of 3 orders of magnitude.
![]()

As a Python user, I utilise excel files to load/store information as business people similar to share data in excel or csv format. Unfortunately, Python is especially boring with Excel files.
In this article, I'll prove you lot five ways to load data in Python. In the end, nosotros'll attain a speedup of 3 orders of magnitude. It'll be lightning-fast.
Edit (xviii/07/2021): I institute a fashion to brand the process 5 times faster (resulting in a 5000x speedup). I added it as a bonus at the end of the article.
Experimental Setup
Allow's imagine that we want to load 10 Exc due east l files with 20000 rows and 25 columns (that'due south around 70MB in total). This is a representative case where yous want to load transactional data from an ERP (SAP) to Python to perform some analysis.
Allow's populate this dummy information and import the required libraries (we'll talk over pickle and joblib later in the article).
import pandas as pd
import numpy as np
from joblib import Parallel, delayed
import fourth dimension for file_number in range(10):
values = np.random.uniform(size=(20000,25))
pd.DataFrame(values).to_csv(f"Dummy {file_number}.csv")
pd.DataFrame(values).to_excel(f"Dummy {file_number}.xlsx")
pd.DataFrame(values).to_pickle(f"Dummy {file_number}.pickle")
five Ways to Load Information in Python
Idea #1: Load an Excel File in Python
Let'south commencement with a straightforward way to load these files. We'll create a kickoff Pandas Dataframe and then append each Excel file to it.
offset = time.time()
df = pd.read_excel("Dummy 0.xlsx")
for file_number in range(1,10):
df.append(pd.read_excel(f"Dummy {file_number}.xlsx"))
end = time.time()
impress("Excel:", cease — start) >> Excel: 53.4
Information technology takes around l seconds to run. Pretty dull.
Idea #2: Use CSVs rather than Excel Files
Let's now imagine that we saved these files as .csv (rather than .xlsx) from our ERP/System/SAP.
start = time.time()
df = pd.read_csv("Dummy 0.csv")
for file_number in range(1,ten):
df.append(pd.read_csv(f"Dummy {file_number}.csv"))
end = time.time()
print("CSV:", cease — offset) >> CSV: 0.632
We tin now load these files in 0.63 seconds. That's most 10 times faster!
Python loads CSV files 100 times faster than Excel files. Use CSVs.
Con: csv files are nearly always bigger than .xlsx files. In this case .csv files are ix.5MB, whereas .xlsx are 6.4MB.
Idea #3: Smarter Pandas DataFrames Creation
We can speed upward our process by changing the way we create our pandas DataFrames. Instead of appending each file to an existing DataFrame,
- We load each DataFrame independently in a list.
- Then concatenate the whole list in a single DataFrame.
offset = time.time()
df = []
for file_number in range(10):
temp = pd.read_csv(f"Dummy {file_number}.csv")
df.append(temp)
df = pd.concat(df, ignore_index=True)
finish = fourth dimension.time()
impress("CSV2:", cease — start) >> CSV2: 0.619
We reduced the time by a few percent. Based on my experience, this trick will become useful when yous deal with bigger Dataframes (df >> 100MB).
Thought #iv: Parallelize CSV Imports with Joblib
Nosotros desire to load ten files in Python. Instead of loading each file i by one, why non loading them all, at once, in parallel?
We can do this hands using joblib.
start = time.time()
def loop(file_number):
return pd.read_csv(f"Dummy {file_number}.csv")
df = Parallel(n_jobs=-1, verbose=10)(delayed(loop)(file_number) for file_number in range(10))
df = pd.concat(df, ignore_index=True)
finish = time.fourth dimension()
print("CSV//:", cease — start) >> CSV//: 0.386
That's most twice as fast as the single cadre version. Still, as a full general dominion, do not await to speed upwards your processes eightfold by using 8 cores (here, I got x2 speed upwards past using 8 cores on a Mac Air using the new M1 chip).
Simple Paralellization in Python with Joblib
Joblib is a unproblematic Python library that allows you to run a office in //. In exercise, joblib works as a list comprehension. Except each iteration is performed by a different thread. Here's an case.
def loop(file_number):
return pd.read_csv(f"Dummy {file_number}.csv")
df = Parallel(n_jobs=-one, verbose=ten)(delayed(loop)(file_number) for file_number in range(10)) #equivalent to
df = [loop(file_number) for file_number in range(ten)]
Thought #5: Utilize Pickle Files
You tin go (much) faster by storing information in pickle files — a specific format used by Python — rather than .csv files.
Con: yous won't be able to manually open a pickle file and see what's in it.
outset = time.time()
def loop(file_number):
render pd.read_pickle(f"Dummy {file_number}.pickle")
df = Parallel(n_jobs=-1, verbose=10)(delayed(loop)(file_number) for file_number in range(10))
df = pd.concat(df, ignore_index=True)
finish = time.time()
print("Pickle//:", finish — start) >> Pickle//: 0.072
We merely cut the running time by 80%!
In general, information technology is much faster to work with pickle files than csv files. Simply, on the other hand, pickles files usually take more infinite on your drive (not in this specific instance).
In practice, y'all will not be able to extract data from a system directly in pickle files.
I would advise using pickles in the ii following cases:
- You want to salvage data from one of your Python processes (and yous don't programme on opening it on Excel) to utilize it after/in some other procedure. Save your Dataframes as pickles instead of .csv
- You need to reload the aforementioned file(s) multiple times. The first time you open up a file, save it every bit a pickle so that you volition be able to load the pickle version direct adjacent fourth dimension.
Example: Imagine that you use transactional monthly information (each month y'all load a new month of data). Yous can salvage all historical information as .pickle and, each time you receive a new file, you tin can load it in one case as a .csv and then keep it every bit a .pickle for the next fourth dimension.
Bonus: Loading Excel Files in Parallel
Permit'south imagine that you received excel files and that you have no other option but to load them as is. You lot can besides employ joblib to parallelize this. Compared to our pickle code from in a higher place, we but demand to update the loop function.
offset = time.time()
def loop(file_number):
return pd.read_excel(f"Dummy {file_number}.xlsx")
df = Parallel(n_jobs=-1, verbose=x)(delayed(loop)(file_number) for file_number in range(ten))
df = pd.concat(df, ignore_index=Truthful)
end = time.fourth dimension()
print("Excel//:", end - start) >> 13.45
Nosotros could reduce the loading time past 70% (from 50 seconds to xiii seconds).
You can also utilise this loop to create pickle files on the wing. So that, next time you load these files, you lot'll be able to accomplish lightning fast loading times.
def loop(file_number):
temp = pd.read_excel(f"Dummy {file_number}.xlsx")
temp.to_pickle(f"Dummy {file_number}.pickle")
return temp Recap
By loading pickle files in parallel, we decreased the loading fourth dimension from 50 seconds to less than a 10th of a 2d.
- Excel: 50 seconds
- CSV: 0.63 seconds
- Smarter CSV: 0.62 seconds
- CSV in //: 0.34 seconds
- Pickle in //: 0.07 seconds
- Excel in //: 13.5 seconds
Bonus #2: 4x Faster Parallelization
Joblib allows to alter the parallelization backend to remove some overheads. Yous can do this by giving adopt="threads" to Parallel.
We obtain a speed of around 0.0096 seconds (over 50 runs with a 2021 MacBook Air).
Using prefer="threads" with CSV and Excel parallelization gives the following results.
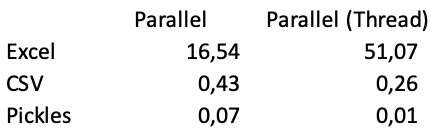
As you lot can see using the "Thread" backend results in a worse score when reading Excel files. But to an astonishing performance with pickles (it takes 50 seconds to load Excel files ane past one, and merely 0.01 seconds to load the data reading pickles files in //).
👉 Allow's connect on LinkedIn!
About the Author
Nicolas Vandeput is a supply chain information scientist specialized in demand forecasting and inventory optimization. He founded his consultancy company SupChains in 2016 and co-founded SKU Scientific discipline — a fast, simple, and affordable demand forecasting platform — in 2018. Passionate about education, Nicolas is both an avid learner and enjoys teaching at universities: he has taught forecasting and inventory optimization to chief students since 2014 in Brussels, Belgium. Since 2020 he is also teaching both subjects at CentraleSupelec, Paris, France. He published Data Science for Supply Chain Forecasting in 2018 (2nd edition in 2021) and Inventory Optimization: Models and Simulations in 2020.

Source: https://towardsdatascience.com/read-excel-files-with-python-1000x-faster-407d07ad0ed8
0 Response to "Python How to Read Data From an Excel"
Post a Comment